Background
Cellular processes are highly dependent on metabolite concentrations and even small perturbations can lead to cell differentiation, apoptosis or cancer.1 A biosensor system that allows determination of metabolite concentrations in vivo is therefore a valuable tool.
An aptamer is a nucleic acid or a peptide molecule that binds to a target with affinities and specificities that rival those of antibodies. Optical aptamers are capable of inducing fluorescence in a target molecule upon binding. An example of this is the RNA aptamer called Spinach (discovered by Paige, et. al in 2011, enhanced by Strack, et. al in 2013).2,3 This aptamer binds the molecule 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) and causes it to fluoresce at a wavelength of 495 nm. It has a fluorescent enhancement that rivals that of green fluorescent protein and it has almost no photobleaching.
In 2012 Paige et. al. found that the Spinach aptamer could be made into a sensor for small molecules by fusing it to other small molecule binding aptamers. In one of the sensors, Spinach was fused with an s-adenosyl methionine (SAM) aptamer (as seen in Figure 1) which made detection of SAM possible.4
This detection was limited to a very small range, which ultimately meant detecting whether or not SAM was present in a reasonable amount - and not the concentration of it. Another issue was that the fluorescence in vivo would not only depend on the concentration of SAM, but also the transcription level of the RNA.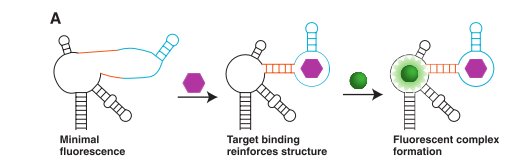
The Project Idea
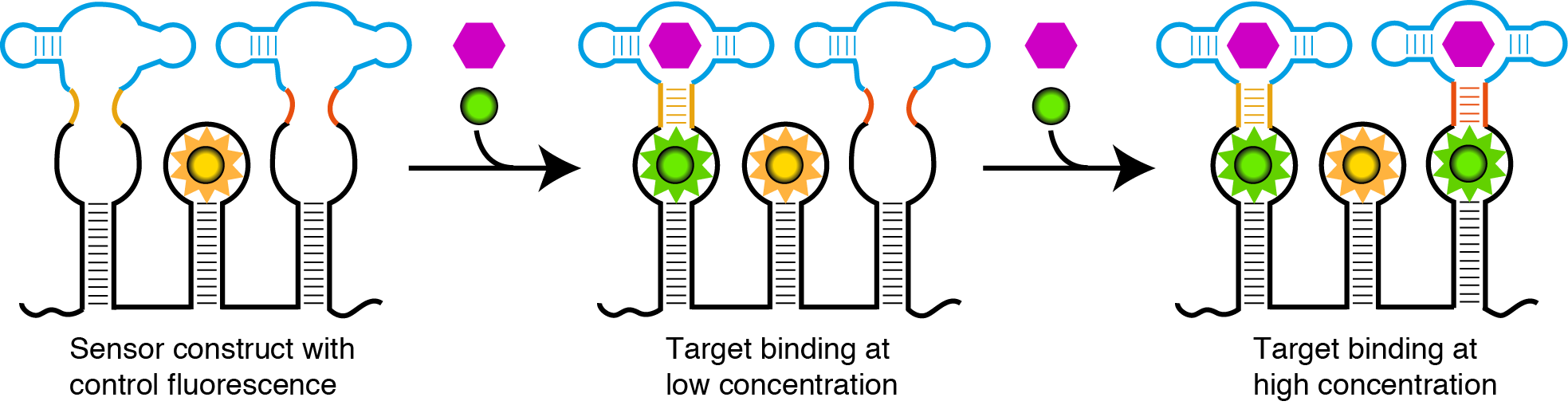
Our idea was to develop an efficient sensor system that can detect small molecules in vivo. We based our sensor on the Spinach-SAM aptamer system described by Paige et. al4 but wanted to extend the concentration range that it can detect. To achieve this we wanted to combine several Spinach-SAM sensors with different affinities for SAM. We reasoned that this could be accomplished by changing the stability of either the linker region between Spinach and SAM aptamers or the SAM detecting aptamer (Fig. 2). Combining the Spinach-SAM aptamers with different affinities for SAM would result in a sensor with a broad dynamic range.5 Next we wanted to add an internal control to determine the concentration of the sensor in relation to the variable SAM dependent signal in vivo. To do this we wanted to include an orthogonal optical aptamer, which would fluoresce independently of the target molecule concentration. The aptamer sensors and control would be integrated on the same RNA strand to obtain known stoichiometry of the sensors and a reliable readout (Fig. 2).
We aim to achieve this by designing different small molecule sensors with different stabilities towards the target molecule and furthermore investigate the orthogonality.
Goals
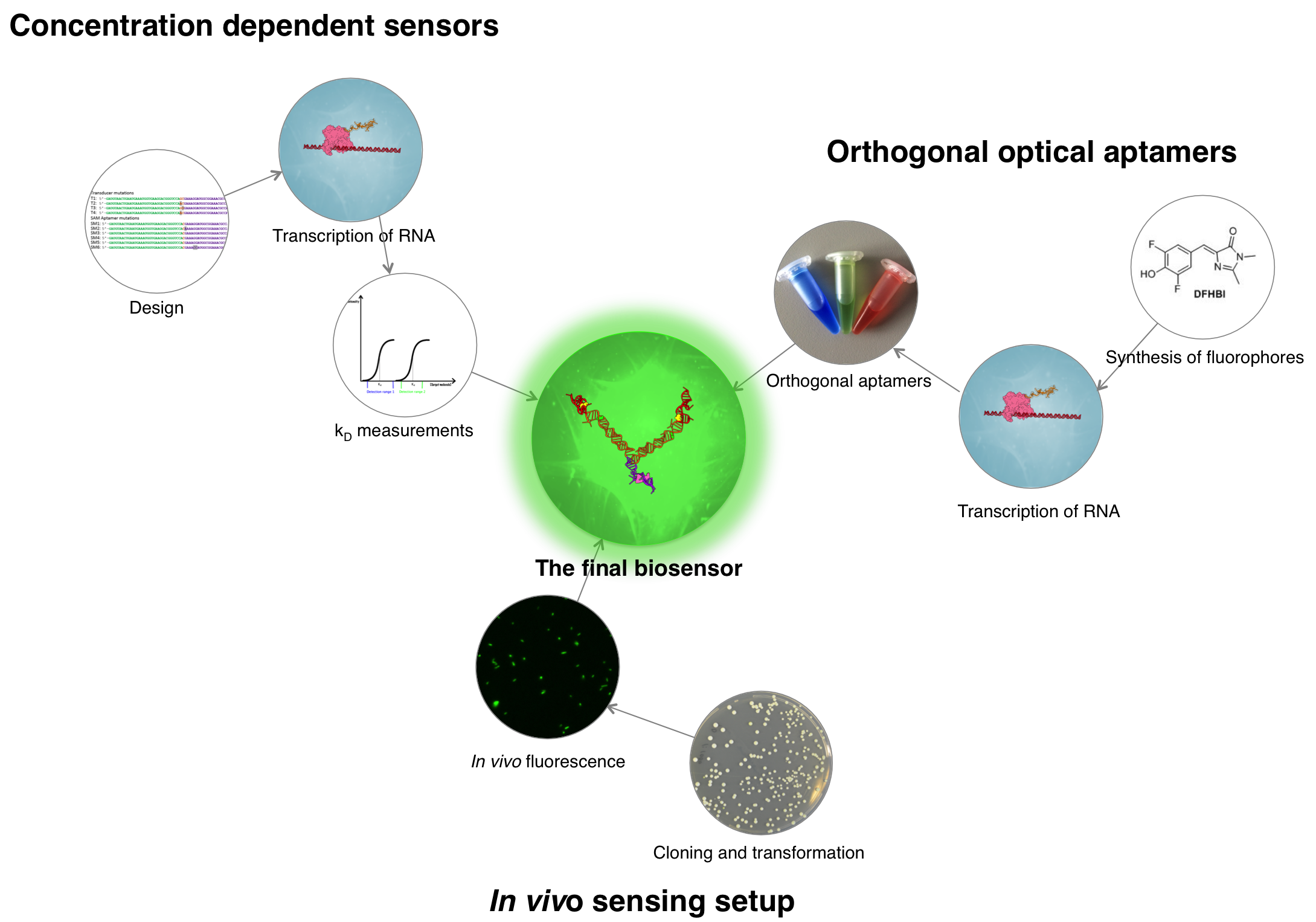
The overall goal of the project was to create a concentration dependent biosensor. The biosensor should consist of multiple small molecule dependent aptamers with different affinities for the target creating a broader dynamic range. To ensure a reliable readout in a cellular setting the sensor must contain an internal control for the transcription level.
In order to achieve this the following subgoals had to be reached:
- Design and test aptamer sensors with different concentration dependency.
- Evaluate fluorophore-aptamer pairs to create an orthogonal internal control for the transcription level.
- Setup an evaluation platform for testing the biosensor in vivo.
We devised the following workflow:
References
- Alberts, B; Johnson, A; Lewis, J; Raff, M; Roberts, K; Walter, P. Molecular Biology of The Cell 5th Edition., Garland Science, Abingdon. p. 1222
- Paige, J. S.; Wu, K. Y.; and Jaffrey, S. R. (2011) RNA mimics of green fluorescent protein. Science, 333, 642.
- Strack, R. L.; Disney, M. D.; and Jaffrey, S. R. (2013) A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods, 10, 1219−1224
- Paige, J. S.; Nguyen-Duc, T.; Song, W.; Jaffrey, S. R. (2012) Fluorescence Imaging of Cellular Metabolites with RNA. Science, 335, 1194
- Vallee-Belisle, A.; Ricci, F.; and Plaxco, K.W. (2012). Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range. J. Am. Chem. Soc. 134, 2876-2879.